GENOmics, Sequencing-based Typing, EpidemioLogy, Linkage, and Antimicrobial Resistance Tool
Powered by high quality data and advanced analytical tools, GENO-STELLARTM is a comprehensive tool that generates insights and predictive models by providing real-time reports with phylogenetic tree mapping and data comparisons to phylogenomic, molecular epidemiology markers, susceptibility data, and clinical outcome information from real-world data samples.
HOW GENO-STELLARTM WORKS
Submit bacterial genome assembly file (FASTA, FA, or FNA file) derived using this pipeline Or other validated assemblers discussed on GENO-STELLARTM’s User Help & Guide. Currently, GENO-STELLARTM tool can only analyze data from highly resistant Klebsiella pneumoniae sensu stricto species.
Evaluate uploaded data against our knowledge base, which houses high quality phylogenomic, clinical, and antibacterial susceptibility data.
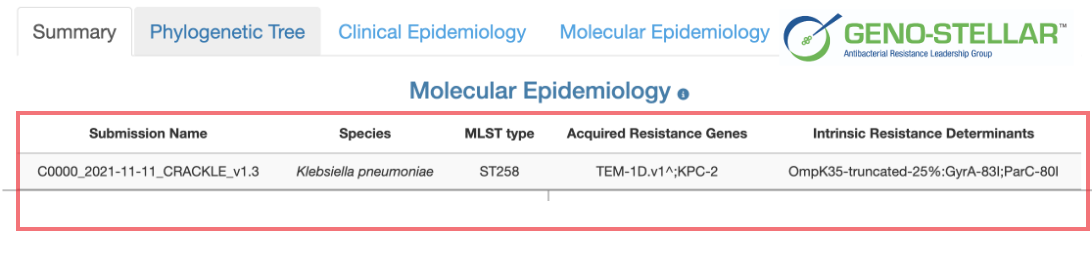
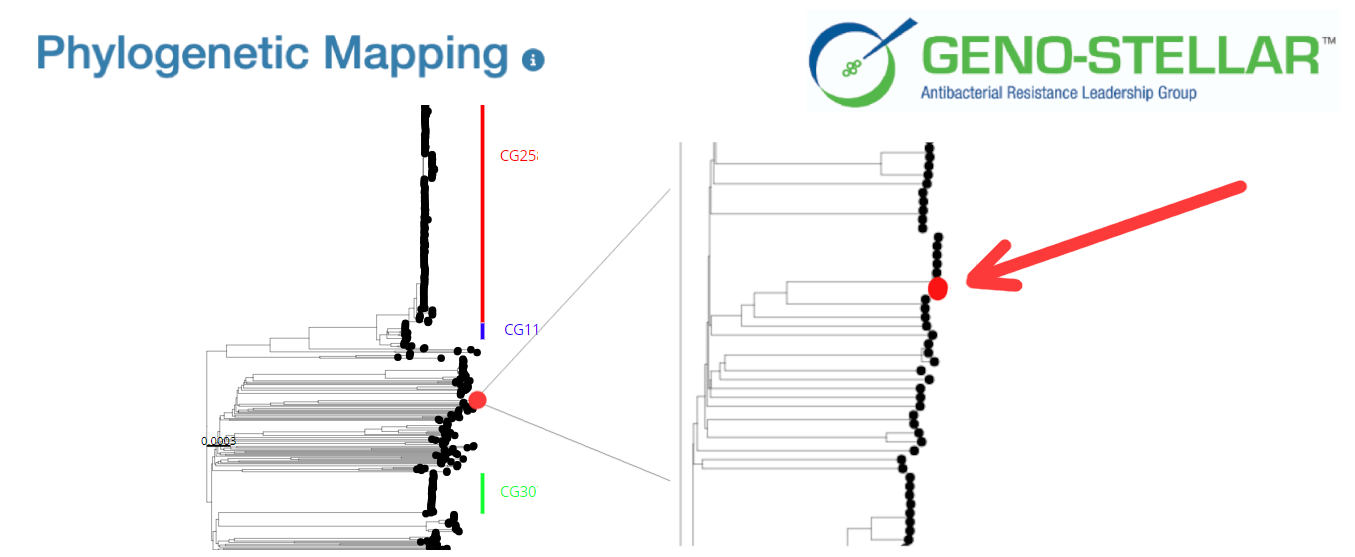
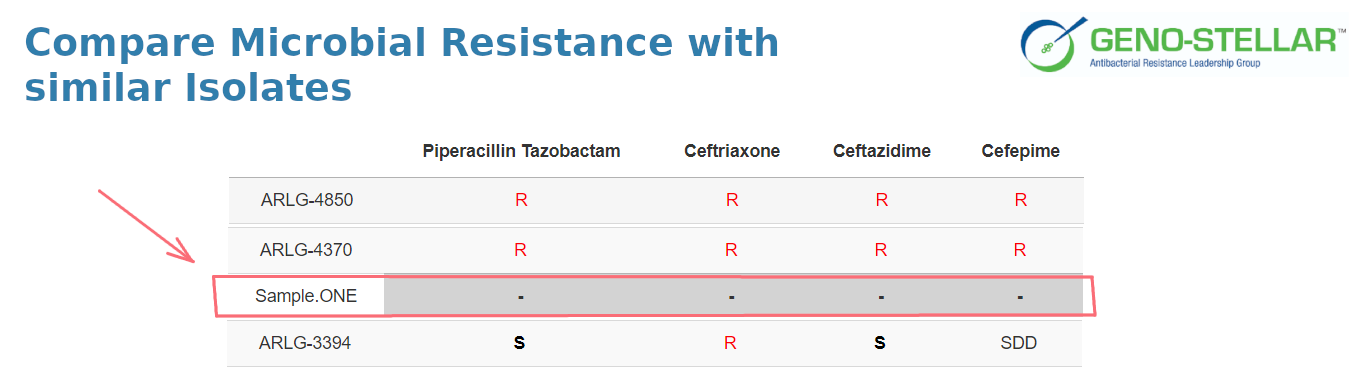
Generate an interactive report describing the molecular epidemiology of the user uploaded isolate, as well as the molecular and clinical features of related isolates from our knowledge base.
OUR MISSION
GENO-STELLARTM was developed through the scientific leadership of the Antibacterial Resistance Leadership Group, which is supported by the National Institute of Allergy and Infectious Diseases under Award Number UM1AI104681.
Our mission with this tool is to incorporate established and cutting-edge sequencing technologies, with advanced analytical tools to generate genomic insights into antibacterial resistance mechanisms in clinical isolates that could aid in the management of patients affected with multi-drug resistant bacteria.
The content of this website is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.